-Search query
-Search result
Showing 1 - 50 of 71 items for (author: lie & tj)
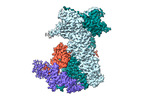
EMDB-18498: 
Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex
Method: single particle / : Kwong HS, Grandvuillemin L, Sirounian S, Ancelin A, Lai-Kee-Him J, Carivenc C, Lancey C, Ragan TJ, Hesketh EL, Bourguet W, Gruszczyk J

EMDB-28850: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab
Method: single particle / : Torres JL, Ward AB

EMDB-28851: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-10 Fab
Method: single particle / : Torres JL, Ward AB

EMDB-28852: 
SARS-CoV-2 Omicron 6P S + COVA309-35 Fab
Method: single particle / : Torres JL, Ward AB
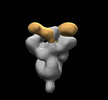
EMDB-28853: 
SARS-CoV-2 Gamma 6P Mut7 + S COVA309-38 Fab
Method: single particle / : Torres JL, Ward AB

EMDB-15786: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form)
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-15787: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-15788: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant)
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-15789: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-15790: 
Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-15791: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0k: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form)
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0l: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0m: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant)
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0n: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0o: 
Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

PDB-8b0p: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3
Method: single particle / : Degtjarik O, Smithers L, Boland C, Caffrey M, Shalev Benami M

EMDB-25100: 
Unmethylated Mtb Ribosome 50S with SEQ-9
Method: single particle / : Xing Z, Cui Z, Zhang J, TB Structural Genomics Consortium (TBSGC)
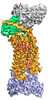
EMDB-25171: 
Cryo-EM structure of ACKR3 in complex with CXCL12, an intracellular Fab, and an extracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG

EMDB-25172: 
Cryo-EM structure of ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ, an intracellular Fab, and an extracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
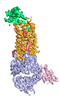
EMDB-25173: 
Cryo-EM structure of ACKR3 in complex with CXCL12 and an intracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
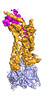
EMDB-25174: 
Cryo-EM structure of human ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ and an intracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
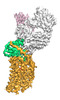
EMDB-25175: 
Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, and an extracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
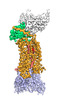
EMDB-25176: 
Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, an extracellular Fab, and an intracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
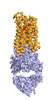
EMDB-25177: 
Cryo-EM structure of human ACKR3 in complex with a small molecule partial agonist CCX662, and an intracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
EMDB-26217: 
Negative stain EM map of COVA1-07 mAb bound to the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB
EMDB-26218: 
Negative stain EM map of COVA2-14 mAb bound to the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB

EMDB-26219: 
Negative stain EM map of COVA2-18 mAb bound to the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB
EMDB-26220: 
Negative stain EM map of the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB

EMDB-22865: 
CryoEM structure of A2296-methylated Mycobacterium tuberculosis ribosome bound with SEQ-9
Method: single particle / : Cui Z, Zhang J

EMDB-25792: 
Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-23914: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-23915: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-23498: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody A23-58.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky Y

EMDB-23499: 
Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody A23-58.1 that targets the receptor-binding domain
Method: single particle / : Zhou T, Tsybovsky T

EMDB-10223: 
Cryo-EM Structure of T. kodakarensis 70S ribosome
Method: single particle / : Matzov D, Sas-Chen A

EMDB-10224: 
Cryo-EM Structure of T. kodakarensis 70S ribosome in TkNat10 deleted strain
Method: single particle / : Matzov D, Sas-Chen A

EMDB-10503: 
Cryo-EM Structure of T. kodakarensis 70S ribosome
Method: single particle / : Matzov D, Sas-Chen A
EMDB-22061: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-15 Fab
Method: single particle / : Ward AB, Bangaru S, Torres JL

EMDB-22062: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA1-22
Method: single particle / : Ward AB, Bangaru S, Torres JL

EMDB-22063: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-07
Method: single particle / : Ward AB, Bangaru S, Torres JL

EMDB-22064: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-39 Fab
Method: single particle / : Ward AB, Bangaru S, Torres JL

EMDB-22065: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-04 Fab
Method: single particle / : Ward AB, Bangaru S, Torres JL

EMDB-22066: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA1-12 Fab
Method: single particle / : Ward AB, Bangaru S, Torres JL

EMDB-4584: 
Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel J, Wollweber F, Pfitzner A, Muehleip A, Sanchez R, Kudryashev M, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M, Kuehlbrandt W, Daumke O
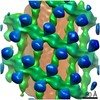
EMDB-10062: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O
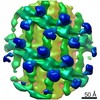
EMDB-10063: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10064: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10065: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzt: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
